Single Cell Analysis Has Been A Mainstay Of Biological Research For Almost 4 Decades. Is It Time To Study Cells On The Basis Of Their Physical Properties, Instead Of Surface Markers?
A Brief History Of Flow Cytometry
Going all the way back to 1879, the physics of droplet formation was explored by Lord Rayleigh, who discovered a property of fluids emerging from a jet orifice is hydrodynamically unstable, and breaks into smaller droplets due to lower surface tension. (Here’s a link to the publication, which is remarkable that you can see what the Royal Society of London’s publications were like almost 150 years ago.)
In 1953, Wallace Coulter received a patent for “Means for Counting Particles Suspended in a Fluid”, where a particle passing through a constricted path will have a detectable change in electrical characteristics. This ‘Coulter Principle’ is used in hemocytometers today to get RBC (Red Blood Cell) counts, a mainstay of blood-based diagnostics. That same year, the principles of laminar flow was used to design a chamber for optical counting of RBCs; it would involve two streams of fluid, a fast-flowing ‘sheath’ and the RBCs injected interior to that stream, where the particles could be measured optically.
This principle of ‘hydrodynamic flow’ is a first key principle of how flow cytometers work today.
At Stanford University in 1965, an advance in ink jet technology used the property of independently-charged droplets for printing applications using a high-speed oscillograph. Thus droplets could be directionally controlled by an electrostatic charge. That same year at Los Alamos National Laboratories, Mack Fulwyler adapted a Coulter Cell Counting instrument to form droplets after counting; these droplets were then charged and deflected into a collection vessel. The principle and construction of the first cell sorting device was reported in this 1965 paper, “Electronic Separation of Biological Cells by Volume”.
The first flow cytometers and cell sorters were commercialized in the late 1960s, and Beckton Dickinson introduced their first instruments in the early 1970’s. A workhorse in the biological sciences, the flow cytometry business (in the aggregate) has flourished, with current revenues above $4 Billion yearly. Applications span both basic research and applied diagnostic testing for cell counting, biomarker detection and cell sorting. The numbers of lasers (and dye combinations) for detection of different markers continue to rise, with as many as 12 fluorescence and 2 scatter parameters measured simultaneously.
Imaging Flow Cytometry (IFC), introduced in 2011 and mainly used to detect extracellular vesicles and circulating exosomes, have increasingly been used for Circulating Tumor Cell (CTC) detection and characterization, as well as cells in states of transition (cell cycle phases such as mitoses). Deepcell has made its first instrument for high throughput imaging of cells only, without any cell labeling to sort cells, but rather their visual properties (that is, their morphology).
High-Speed Brightfield Imaging From Deepcell
Deepcell was co-founded in the laboratory of Euan Ashley at Stanford University with Maddison Masaeli, who was a postdoc in the Ashley lab, and Mahyar Salek, who was involved in deep learning at both Google and Emotiful, with the concept to combine high-resolution imaging of single cells with the sorting of these cells in a label-free fashion by leveraging real-time artificial intelligence and deep learning.
In May of 2023 the benchtop REM-I platform was announced. This platform is able to image cells at high speed, classify their morphology with Deepcell’s Human Foundation Model, and then sort them without the use of labels. Deepcell’s Axon data suite enables the user to represent the 115 dimensions of morphology measured by the Human Foundation Model as UMAP (2D Uniform Manifold Approximation and Dimensions) plots, a convenient method to reduce high dimensional data into a 2-dimensional map. (See figure 1 for a simple example of three cell lines mixed with polystyrene beads run on the REM-I platform).
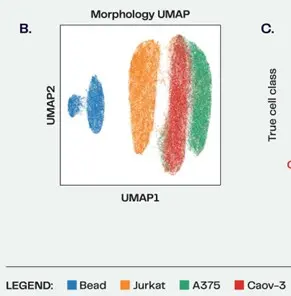
Figure 1: Evaluation of the Deepcell HFM with B) polystyrene beads and three cell lines. 6μm polystyrene beads, cancer (A375 and Caov-3) and immune (Jurkat) cell lines were imaged on the Deepcell REM-I platform, then combined in silico to evaluate the classification performance of the HFM.
Real-Time AI Characterization In 115 Dimensions
Single cell analysis has progressed from a light microscope to genomic characterization of a cell population via single cell transcriptome analysis of all the expressed genes in the genome. The REM-I instrument takes cells in suspension, and are loaded and analyzed by an automated microfluidic system. The instrument captures high-resolution brightfield images, and can sort cells in six different ways for downstream analysis. (A typical analysis would be single cell RNA-Seq whole transcriptome analysis.) See figure 2 for an illustration of how the instrument works.
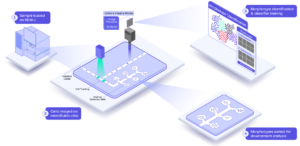
Figure 2: The REM-I Instrument workflow, from cell suspension through examination of high-dimension cell morphology analysis, selecting cells of interest and recovering sorted cells.
Through an artificial intelligence data model Deepcell calls the Human Foundation Model, cells can be identified through computer vision (to measure morphometrics) and self-supervised deep learning to derive 115 dimensions from each image. There are two different encoders analyzing an image: one called the Deep Learning Encoder (utilizing a Convolutional Neural Network) that extracts 64 dimensions of Deep Learning features, and a second one called the Computer Vision Encoder (utilizing human constructed algorithms) extracting an additional 51 dimensions of morphometric features. These 51 morphometric dimensions are what you normally consider as morphology – cell shape, pixel intensity, and image texture, while the 64 Deep Learning dimensions are not human-interpretable.
Their Human Foundation Model has been pre-trained on a variety of cell types, including normal adult blood cells, fetal blood, trophoblast cell lines and a variety of cancer cell lines. Over 25 million high-resolution images were used to derive AI-assisted cell image annotation.
It’s remarkable that so many parameters could be extracted from a single brightfield image, and in real-time; in a 2023 Nature publication (Communications Biology), a paper about the REM-I platform technology (“COSMOS: a platform for real-time morphology-based, label-free cell sorting using deep learning”) showed data from 600 cells per second up to 6,000 cells per second.
Will Morpholomics Become Mainstream?
It should come as no surprise that with the wealth of genomic, transcriptomic, epigenetic and proteomic datasets now available, there is great interest at the intersection of these “omics” for furthering both basic research and personalized medicine. Other less-known fields such as metabolomics and lipidomics are limited through current methods of analysis, and may well become more important in the future as better methods of analyses become available.
At the October 2023 American Society for Human Genetics conference in Washington DC, Deepcell presented a poster called “Morphology profiling driven by deep learning characterizes functional changes in CRISPR knockout cell lines” illustrating how morphology changes with single gene knockouts. 11 genes were individually down-regulated using CRISPR, and it was clear that distinct morphotypes could be distinguished between the individual edited cell lines. The cell images in the poster and its relationship to the respective area on the UMAP plots were convincing. (This poster is available for download from the Deepcell website here (PDF).)
Could the “morpholome” become an accepted area of study? One tantalizing application is in the realm of non-invasive prenatal diagnostics; Deepcell’s CEO Dr. Maddison Masaeli said in this 2023 interview that circulating Trisomy 21 fetal cells were different than normal healthy fetal cells, and visibly so. This opens up the possibility for early and non-invasive screening methods for detecting certain genetic conditions. The potential for circulating tumor cell (CTC) detection is obvious, which has been intensively studied since the advent of the cell-surface marker EpCAM was used in the CellSearch CTC platform.[1]
Other application areas Deepcell is targeting include metastatic cancer research, translational research and Alzheimer’s Disease research (several presentation webinars covering these topics are on Deepcell’s Webinars page)
There are several imaging flow cytometers on the market, however none with the deep learning cell model and real-time sorting capabilities in a label-free manner that Deepcell offers. [2] BioCompare has a brief review called Optimizing Cell Sorting covering the challenges of fluorescent probe panel design to look into this issue in a little more detail.
Availability Of Datasets And Ability To Access Technology
If you’d like to take a look at several datasets along with Deepcell’s Axon software, you can sign up here or you can request a personal demo. Deepcell also offers a technology access capability they call the Spark Program, where you can send your samples to Deepcell for analysis, available here.
Reference:
- Salek M and Masaeli et al. (2023) Commun Biol. 6(1):971. COSMOS: a platform for real-time morphology-based, label-free cell sorting using deep learning doi:10.1038/s42003-023-05325-9

Dale Yuzuki
Multiomics Writer



